Stranded RNA-seq - antisense transcription zfCRISPRi
Experimental summary
CRISPRi microinjections were performed with Ac/Ds-sgRNAs targeting initiation of antisense transcription at sox9a and foxd3 loci. Scrambled guides were used as control. Cells expressing dCas9-SID4x-2a-Citrine under the control of a sox10 (most neural crest, and few non-NC cell types) BAC transgene were FAC-sorted from 24hpf embryos. RNA were isolated for strand-specific RNAseq library preparation using dUTP protocol (KAPA RNA HyperPrep Kit with RiboErase (HMR)). Transcript quantification followed by differential expression was performed using kallisto/sleuth pipeline.
Key experimental protocols can be found here.
Tarball (25Gb) containing FastQ files and custom sequences can be downloaded from here.
Transcript quantification
Quality of reads were checked with FastQC v0.11.9 prior to following analysis.
Trim reads - cutadapt v2.10 (Python 3.6.10)
Libraries were indexed using NEBNext adapters and oligos.
cutadapt -a Read1_F=AGATCGGAAGAGCACACGTCTGAACTCCAGTCA -A Read2_R=AGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGT \
-m 40 -q 30 --cores=8 --report=minimal \
-o trimmed_cit_scr-1_R1.fastq -p trimmed_cit_scr-1_R2.fastq \
cit_scr-1_R1.fastq cit_scr-1_R2.fastqTrimming of adapters from reads were confirmed by rerunning FastQC v0.11.9.
Transcriptome reference for pseudoalignment
Download Ensembl release 105 sequences and chrom sizes from UCSC Genome Browser.
wget http://ftp.ensembl.org/pub/release-105/fasta/danio_rerio/cdna/Danio_rerio.GRCz11.cdna.all.fa.gz
wget http://ftp.ensembl.org/pub/release-105/fasta/danio_rerio/ncrna/Danio_rerio.GRCz11.ncrna.fa.gz
wget http://ftp.ensembl.org/pub/release-105/gtf/danio_rerio/Danio_rerio.GRCz11.105.gtf.gz
wget https://hgdownload.soe.ucsc.edu/goldenPath/danRer11/bigZips/danRer11.chrom.sizesCreate custom transcriptome reference combining cDNA (Ensembl), ncRNA (Ensembl), sox9a_AS (custom), and foxd3_AS (custom).
cat Danio_rerio.GRCz11.cdna.all.fa Danio_rerio.GRCz11.ncrna.fa sox9a_AS.fa foxd3_AS.fa > custom_14Mar2022.faIndex for pseudoalignment - kallisto v0.46.1
kallisto index -i custom_14Mar2022.idx custom_14Mar2022.fa --make-unique#[build] loading fasta file custom_14Mar2022.fa
#[build] k-mer length: 31
#[build] warning: clipped off poly-A tail (longer than 10) from 380 target sequences
#[build] warning: replaced 13167 non-ACGUT characters in the input sequence with pseudorandom nucleotides
#[build] counting k-mers ... done.
#[build] building target de Bruijn graph ... done
#[build] creating equivalence classes ... done
#[build] target de Bruijn graph has 501714 contigs and contains 71214712 k-mersQuantify transcripts - kallisto v0.46.1
Include bootstrapping option for sleuth differential expression. Include –genomebam option to project pseudoalignments onto genome sorted BAM files.
kallisto quant -i ensembl_105/custom_14Mar2022.idx \
-o kallisto/cit_scr-1 \
-b 100 --genomebam -g ensembl_105/Danio_rerio.GRCz11.105.gtf -c danRer11.chrom.sizes \
fastq/trimmed_cit_scr-1_R1.fastq fastq/trimmed_cit_scr-1_R2.fastq \
--rf-stranded --threads=8Differential transcript expression and GO analysis
Load required packages
library(sleuth)
library(tidyverse)## ── Attaching packages ─────────────────────────────────────── tidyverse 1.3.2 ──
## ✔ ggplot2 3.3.6 ✔ purrr 0.3.5
## ✔ tibble 3.1.8 ✔ dplyr 1.0.10
## ✔ tidyr 1.2.1 ✔ stringr 1.4.1
## ✔ readr 2.1.3 ✔ forcats 0.5.2
## ── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
## ✖ dplyr::filter() masks stats::filter()
## ✖ dplyr::lag() masks stats::lag()library(biomaRt)
library(rbioapi)Load kallisto data and assign experimental setup
sample_id <- dir(file.path(".", "abundances"))
kal_dirs <- file.path(".", "abundances", sample_id)
s2c <- read.table(file.path(".", "seq_info.txt"), header = TRUE)
s2c <- dplyr::select(s2c, sample = sample, condition)
s2c <- dplyr::mutate(s2c, path = kal_dirs)
s2c$condition <- relevel(factor(s2c$condition), "scr") # set "scr" as the base level (! IMPORTANT !)Collect Ensembl gene names using biomaRt
mart <- biomaRt::useMart(biomart = "ENSEMBL_MART_ENSEMBL", dataset = "drerio_gene_ensembl", host = 'https://www.ensembl.org')
t2g <- biomaRt::getBM(attributes = c("ensembl_transcript_id_version", "ensembl_gene_id", "external_gene_name", "description"), mart = mart)
t2g <- dplyr::rename(t2g, target_id = ensembl_transcript_id_version, ens_gene = ensembl_gene_id, ext_gene = external_gene_name, des=description)Differential transcript expression - sleuth v0.30.0
- Construct “sleuth object”. Stores information about the experiment, and details of the model to be used for differential testing.
- Fit data to models for hypothesis testing.
- “Smooth” the raw kallisto abundance estimates for each sample using a linear model with a parameter that represents the experimental condition.
- Perform Wald test.
set.seed(584)
so <- sleuth_prep(s2c, extra_bootstrap_summary = TRUE, read_bootstrap_tpm = TRUE,
target_mapping = t2g, transformation_function = function(x) log2(x + 0.01))## Warning in check_num_cores(num_cores): It appears that you are running Sleuth from within Rstudio.
## Because of concerns with forking processes from a GUI, 'num_cores' is being set to 1.
## If you wish to take advantage of multiple cores, please consider running sleuth from the command line.## reading in kallisto results## dropping unused factor levels## ....
## normalizing est_counts
## 41561 targets passed the filter
## normalizing tpm
## merging in metadata
## summarizing bootstraps
## ....so <- sleuth_fit(so, ~condition, 'full')## fitting measurement error models
## shrinkage estimation
## 56 NA values were found during variance shrinkage estimation due to mean observation values outside of the range used for the LOESS fit.
## The LOESS fit will be repeated using exact computation of the fitted surface to extrapolate the missing values.
## These are the target ids with NA values: ENSDART00000076321.5, ENSDART00000084355.5, ENSDART00000097712.5, ENSDART00000112372.5, ENSDART00000115803.2, ENSDART00000118229.2, ENSDART00000122139.3, ENSDART00000122387.3, ENSDART00000127455.3, ENSDART00000128551.4, ENSDART00000133528.3, ENSDART00000134971.3, ENSDART00000135183.2, ENSDART00000137700.2, ENSDART00000138799.2, ENSDART00000141683.2, ENSDART00000141932.2, ENSDART00000143741.2, ENSDART00000144348.3, ENSDART00000147102.2, ENSDART00000147419.2, ENSDART00000153466.3, ENSDART00000153805.2, ENSDART00000153994.2, ENSDART00000154956.2, ENSDART00000156029.3, ENSDART00000156122.2, ENSDART00000160217.2, ENSDART00000162455.2, ENSDART00000164803.2, ENSDART00000166721.2, ENSDART00000167092.2, ENSDART00000167288.2, ENSDART00000169153.2, ENSDART00000172243.2, ENSDART00000173588.2, ENSDART00000177618.2, ENSDART00000178367.2, ENSDART00000178620.2, ENSDART00000180676.1, ENSDART00000182261.1, ENSDART00000182989.1, ENSDART00000185294.1, ENSDART00000186413.1, ENSDART00000188517.1, ENSDART00000188888.1, ENSDART00000190726.1, ENSDART00000191642.1, ENSDART00000193182.1, ENSDART00000194154.1, ENSDART00000194196.1, ENSDART00000194317.1, ENSDART00000023156.7, ENSDART00000093606.3, ENSDART00000163316.2, ENSDART00000168497.2
## computing variance of betasso <- sleuth_wt(so, 'conditionlnc', which_model="full") # Wald test
sleuth_table <- sleuth_results(so, "conditionlnc", "full", show_all = TRUE) # Examine results
# uncomment to save results locally
#write.csv(sleuth_table,file='wald_conditionlnc.csv')
#save(so, file='wald_conditionlnc.RData')Plots to visualise samples.
plot_pca(so, color_by = 'condition', text_labels = TRUE) + theme_minimal()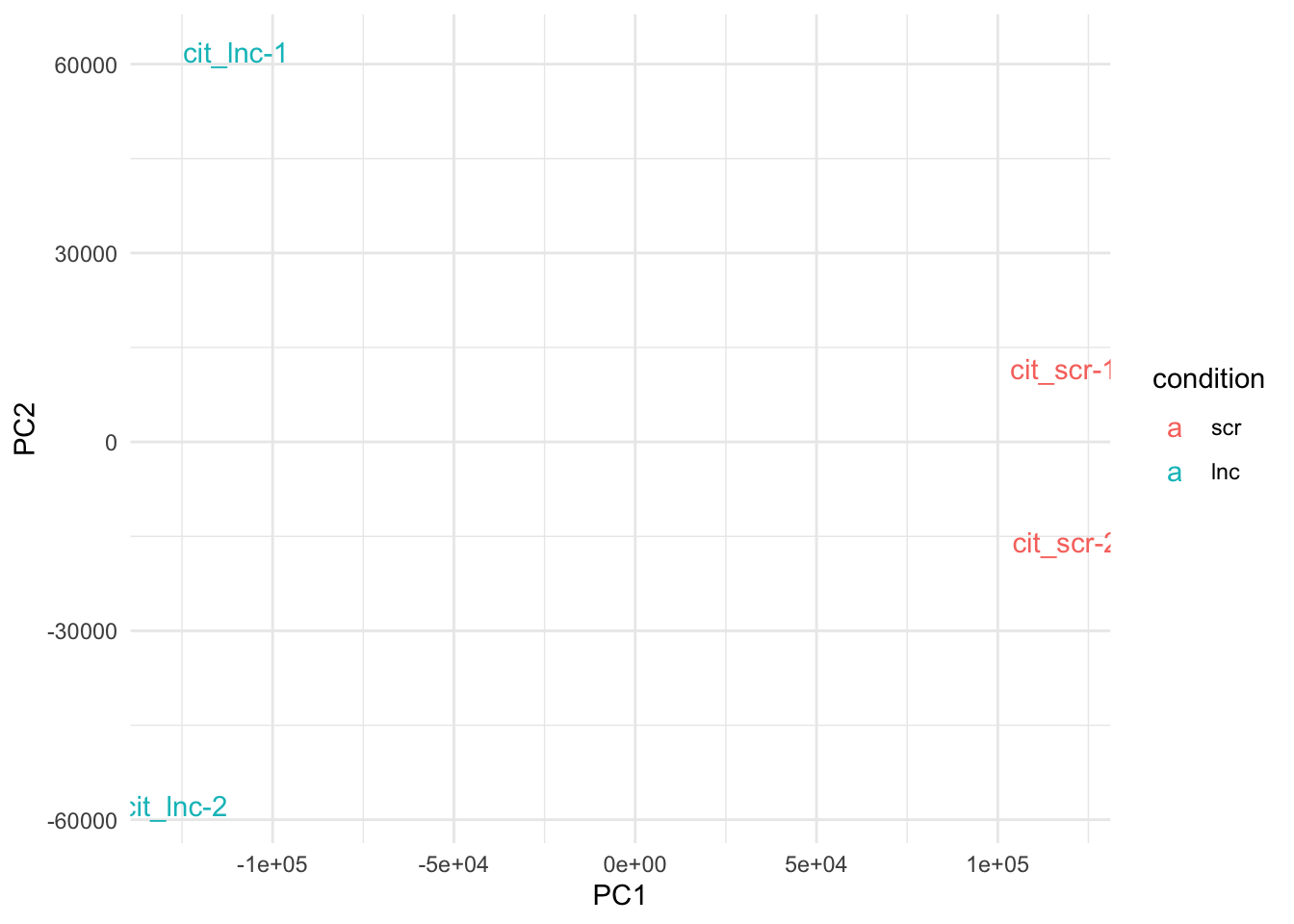
plot_sample_heatmap(so)Result tables.
sig_transcripts_up <- sleuth_table %>% filter(qval < 0.05, b>0)
sig_transcripts_dn <- sleuth_table %>% filter(qval < 0.05, b<0)
# uncomment to save results locally
#write.csv(sig_transcripts_up,file='wald_conditionlnc_up_qval005.csv')
#write.csv(sig_transcripts_dn,file='wald_conditionlnc_down_qval005.csv')Bootstrap estimates of sox9a/foxd3 transcripts.
plot_bootstrap(so,
target_id = "sox9a_AS",
units = "tpm",
color_by = "condition") + theme_minimal()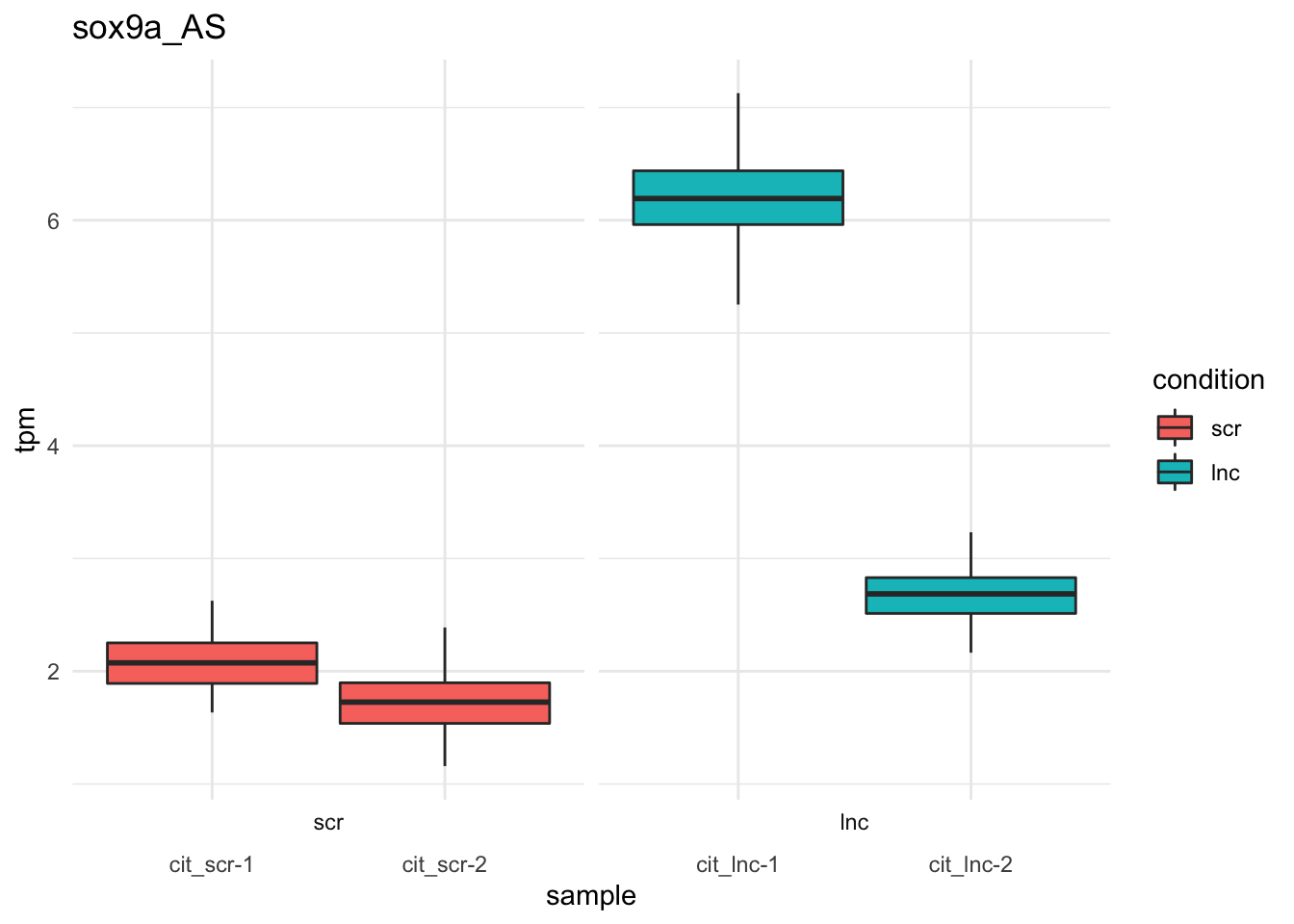
plot_bootstrap(so,
target_id = "foxd3_AS",
units = "tpm",
color_by = "condition") + theme_minimal()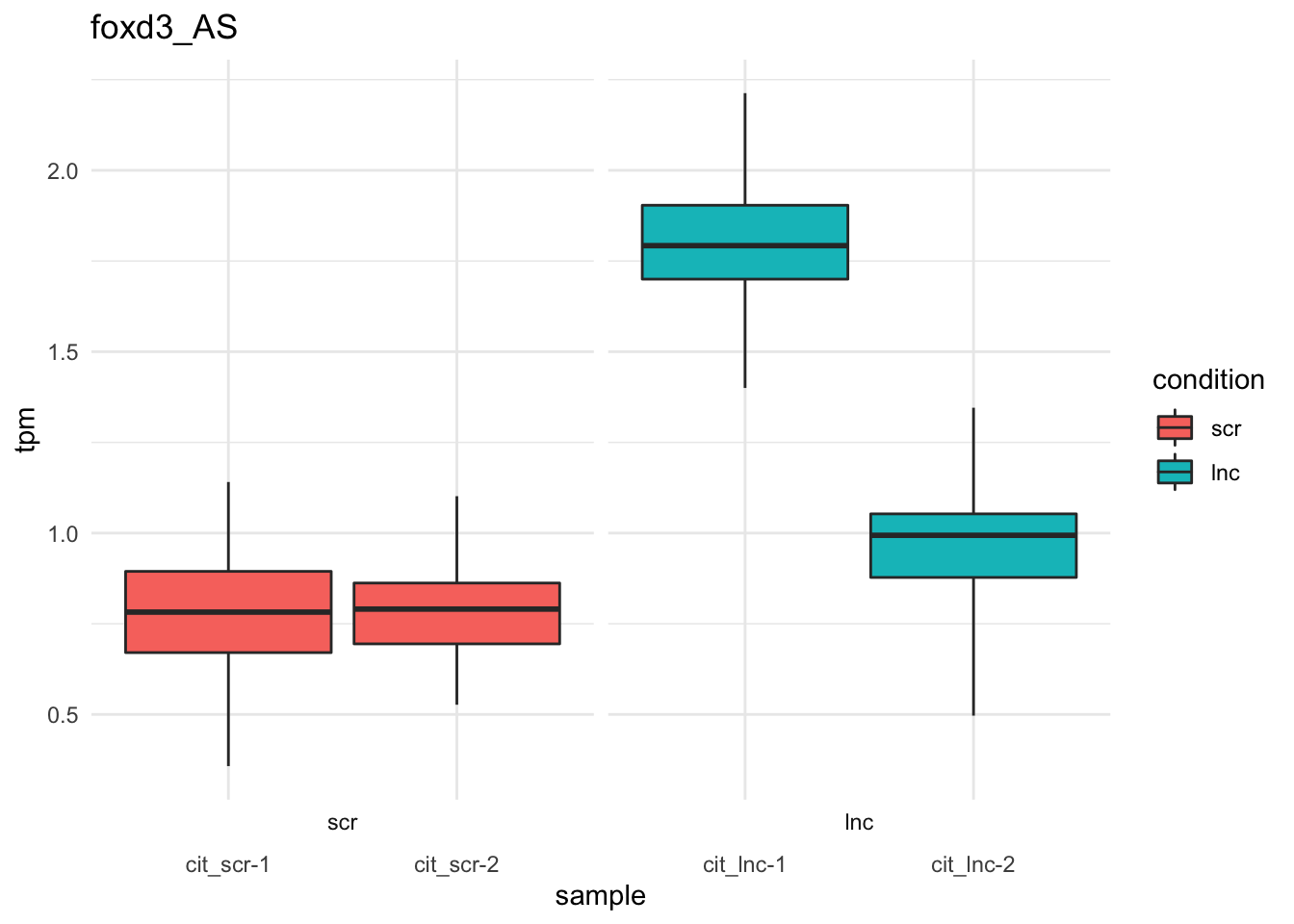
plot_bootstrap(so,
target_id = "ENSDART00000005676.6", # sox9a-201
units = "tpm",
color_by = "condition") + theme_minimal()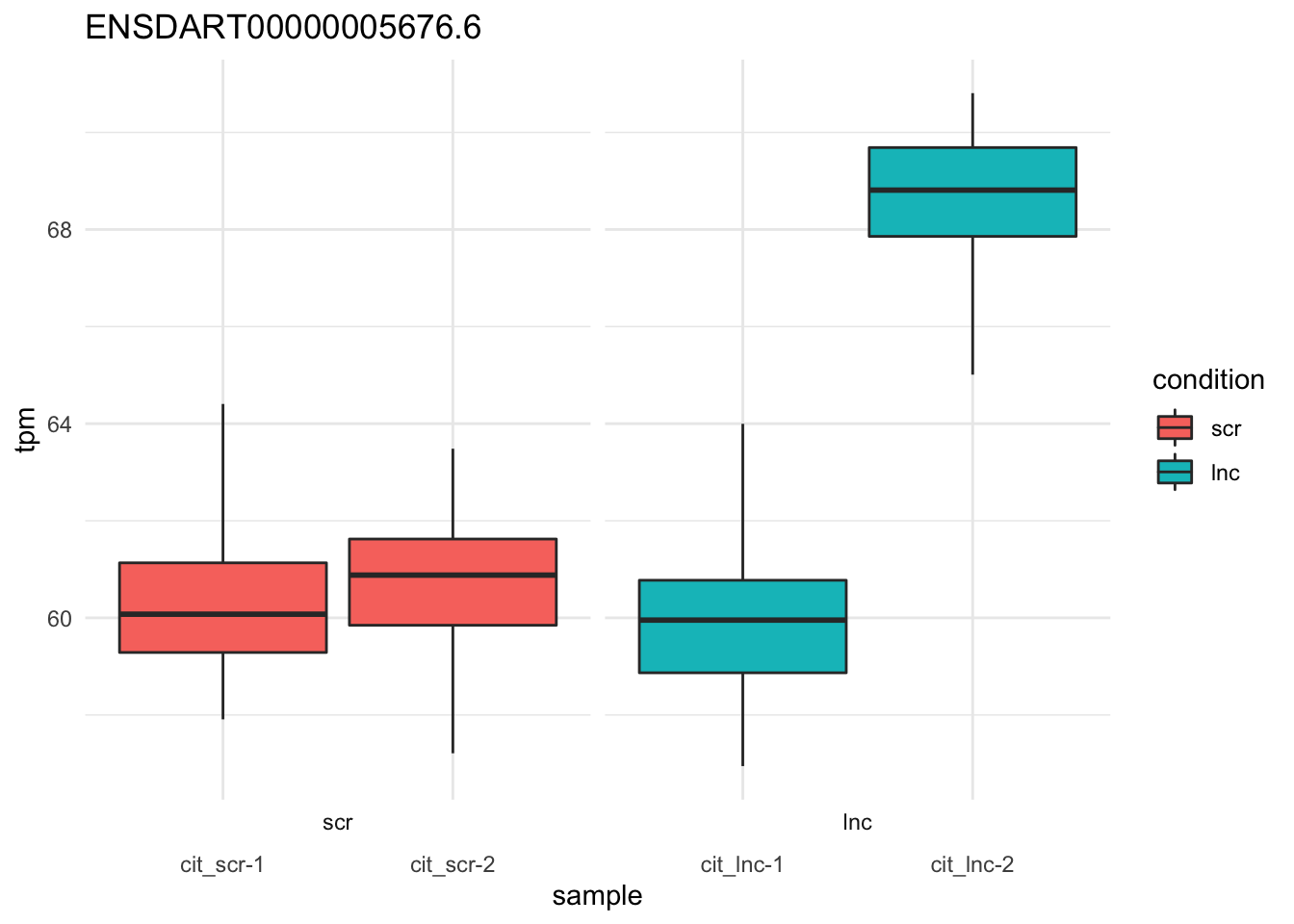
plot_bootstrap(so,
target_id = "ENSDART00000127937.3", # sox9a-202
units = "tpm",
color_by = "condition") + theme_minimal()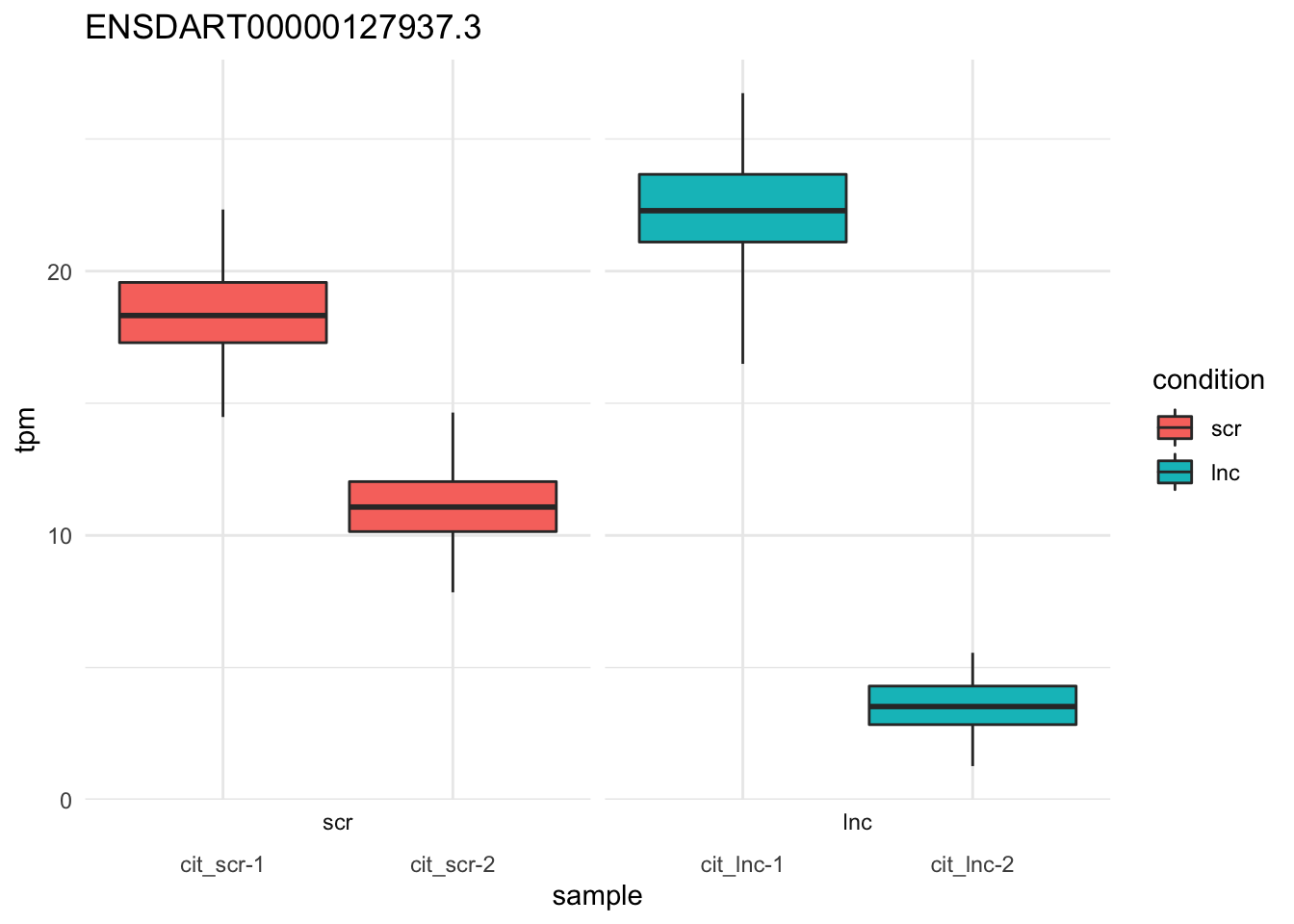
plot_bootstrap(so,
target_id = "ENSDART00000017695.6", # foxd3-201
units = "tpm",
color_by = "condition") + theme_minimal()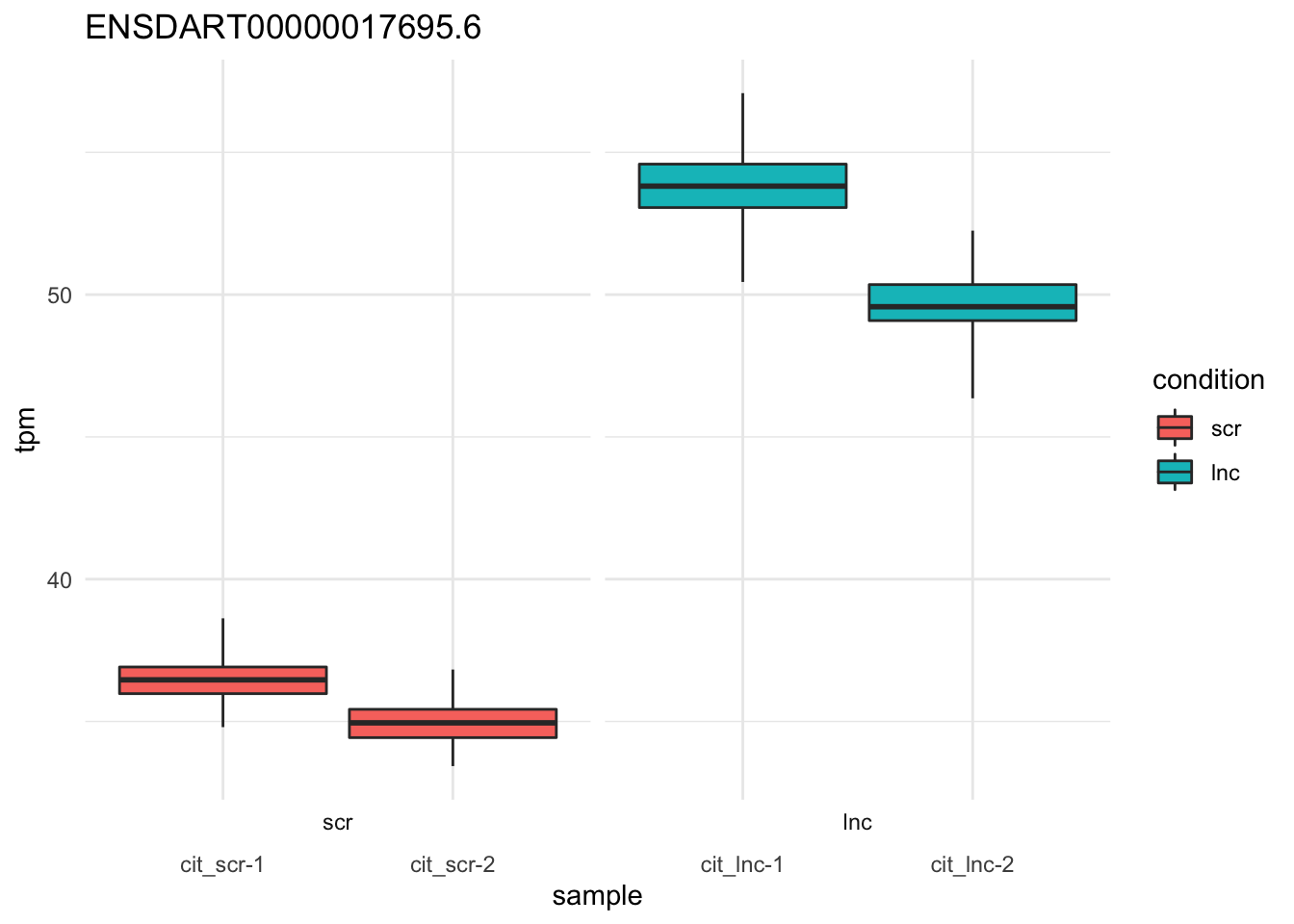
GO analysis - PANTHER classification system
up_genes <- as.character(sig_transcripts_up$ens_gene)
down_genes <- as.character(sig_transcripts_dn$ens_gene)
data.frame(rba_panther_info(what="datasets")) # to choose dataset ID## Retrieving available annotation datasets.## release_date
## 1 2022-07-01
## 2 2022-07-01
## 3 2022-07-01
## 4 2022-02-22
## 5 2022-02-22
## 6 2022-02-22
## 7 2022-02-22
## 8 2022-02-22
## 9 2021-10-01
## description
## 1 Gene Ontology Molecular Function annotations including both manually curated and electronic annotations.
## 2 Gene Ontology Biological Process annotations including both manually curated and electronic annotations.
## 3 Gene Ontology Cellular Component annotations including both manually curated and electronic annotations.
## 4 A molecular process that can be carried out by the action of a single macromolecular machine, usually via direct physical interactions with other molecular entities. Function in this sense denotes an action, or activity, that a gene product (or a complex) performs. These actions are described from two distinct but related perspectives: (1) biochemical activity, and (2) role as a component in a larger system/process.
## 5 A biological process represents a specific objective that the organism is genetically programmed to achieve. Biological processes are often described by their outcome or ending state, e.g., the biological process of cell division results in the creation of two daughter cells (a divided cell) from a single parent cell. A biological process is accomplished by a particular set of molecular functions carried out by specific gene products (or macromolecular complexes), often in a highly regulated manner and in a particular temporal sequence.
## 6 A location, relative to cellular compartments and structures, occupied by a macromolecular machine when it carries out a molecular function. There are two ways in which the gene ontology describes locations of gene products: (1) relative to cellular structures (e.g., cytoplasmic side of plasma membrane) or compartments (e.g., mitochondrion), and (2) the stable macromolecular complexes of which they are parts (e.g., the ribosome).
## 7
## 8 Panther Pathways
## 9 Reactome Pathways
## id label
## 1 GO:0003674 molecular_function
## 2 GO:0008150 biological_process
## 3 GO:0005575 cellular_component
## 4 ANNOT_TYPE_ID_PANTHER_GO_SLIM_MF PANTHER GO Slim Molecular Function
## 5 ANNOT_TYPE_ID_PANTHER_GO_SLIM_BP PANTHER GO Slim Biological Process
## 6 ANNOT_TYPE_ID_PANTHER_GO_SLIM_CC PANTHER GO Slim Cellular Location
## 7 ANNOT_TYPE_ID_PANTHER_PC protein class
## 8 ANNOT_TYPE_ID_PANTHER_PATHWAY ANNOT_TYPE_PANTHER_PATHWAY
## 9 ANNOT_TYPE_ID_REACTOME_PATHWAY ANNOT_TYPE_REACTOME_PATHWAY
## version
## 1 10.5281/zenodo.6799722
## 2 10.5281/zenodo.6799722
## 3 10.5281/zenodo.6799722
## 4 17
## 5 17
## 6 17
## 7 17
## 8 17
## 9 65data.frame(rba_panther_info(what="organisms")) # to choose organism ID## Retrieving supported organisms in PANTHER.## name taxon_id short_name version
## 1 human 9606 HUMAN Reference Proteome 2021_03
## 2 mouse 10090 MOUSE Reference Proteome 2021_03
## 3 rat 10116 RAT Reference Proteome 2021_03
## 4 chicken 9031 CHICK Reference Proteome 2021_03
## 5 zebrafish 7955 DANRE Reference Proteome 2021_03
## 6 fruit_fly 7227 DROME Reference Proteome 2021_03
## 7 nematode_worm 6239 CAEEL Reference Proteome 2021_03
## 8 budding_yeast 559292 YEAST Reference Proteome 2021_03
## 9 fission_yeast 284812 SCHPO Reference Proteome 2021_03
## 10 dictyostelium 44689 DICDI Reference Proteome 2021_03
## 11 arabidopsis 3702 ARATH Reference Proteome 2021_03
## 12 e_coli 83333 ECOLI Reference Proteome 2021_03
## 13 aspergillus 227321 EMENI Reference Proteome 2021_03
## 14 Amborella 13333 AMBTC Reference Proteome 2021_03
## 15 lizard 28377 ANOCA Reference Proteome 2021_03
## 16 mosquito 7165 ANOGA Reference Proteome 2021_03
## 17 aquifex 224324 AQUAE Reference Proteome 2021_03
## 18 ashbya 284811 ASHGO Reference Proteome 2021_03
## 19 bacillus_cereus 226900 BACCR Reference Proteome 2021_03
## 20 bacillus_subtilis 224308 BACSU Reference Proteome 2021_03
## 21 bacteroidetes 226186 BACTN Reference Proteome 2021_03
## 22 chytrid 684364 BATDJ Reference Proteome 2021_03
## 23 cow 9913 BOVIN Reference Proteome 2021_03
## 24 purple_false_brome 15368 BRADI Reference Proteome 2021_03
## 25 bradyrhizobium 224911 BRADU Reference Proteome 2021_03
## 26 branchiostoma 7739 BRAFL Reference Proteome 2021_03
## 27 canola 3708 BRANA Reference Proteome 2021_03
## 28 cabbage 51351 BRARP Reference Proteome 2021_03
## 29 c_briggsae 6238 CAEBR Reference Proteome 2021_03
## 30 candida 237561 CANAL Reference Proteome 2021_03
## 31 dog 9615 CANLF Reference Proteome 2021_03
## 32 bell pepper 4072 CAPAN Reference Proteome 2021_03
## 33 chlamydia 272561 CHLTR Reference Proteome 2021_03
## 34 green_algae 3055 CHLRE Reference Proteome 2021_03
## 35 chloroflexus 324602 CHLAA Reference Proteome 2021_03
## 36 ciona 7719 CIOIN Reference Proteome 2021_03
## 37 orange 2711 CITSI Reference Proteome 2021_03
## 38 clostridium 441771 CLOBH Reference Proteome 2021_03
## 39 coxiella 227377 COXBU Reference Proteome 2021_03
## 40 cryptococcus 214684 CRYNJ Reference Proteome 2021_03
## 41 cucumber 3659 CUCSA Reference Proteome 2021_03
## 42 water_flea 6669 DAPPU Reference Proteome 2021_03
## 43 deinococcus 243230 DEIRA Reference Proteome 2021_03
## 44 dictyoglomus 515635 DICTD Reference Proteome 2021_03
## 45 d_purpureum 5786 DICPU Reference Proteome 2021_03
## 46 entamoeba 5759 ENTHI Reference Proteome 2021_03
## 47 horse 9796 HORSE Reference Proteome 2021_03
## 48 yellow monkey flower 4155 ERYGU Reference Proteome 2021_03
## 49 flooded gum 71139 EUCGR Reference Proteome 2021_03
## 50 cat 9685 FELCA Reference Proteome 2021_03
## 51 fusobacterium 190304 FUSNN Reference Proteome 2021_03
## 52 geobacter 243231 GEOSL Reference Proteome 2021_03
## 53 giardia 184922 GIAIC Reference Proteome 2021_03
## 54 gloeobacter 251221 GLOVI Reference Proteome 2021_03
## 55 soybean 3847 SOYBN Reference Proteome 2021_03
## 56 gorilla 9595 GORGO Reference Proteome 2021_03
## 57 cotton 3635 GOSHI Reference Proteome 2021_03
## 58 h_flu 71421 HAEIN Reference Proteome 2021_03
## 59 halobacterium 64091 HALSA Reference Proteome 2021_03
## 60 sunflower 4232 HELAN Reference Proteome 2021_03
## 61 h_pylori 85962 HELPY Reference Proteome 2021_03
## 62 barley 112509 HORVV Reference Proteome 2021_03
## 63 tick 6945 IXOSC Reference Proteome 2021_03
## 64 english walnut 51240 JUGRE Reference Proteome 2021_03
## 65 green algae 105231 KLENI Reference Proteome 2021_03
## 66 korarchaeum 374847 KORCO Reference Proteome 2021_03
## 67 garden lettuce 4236 LACSA Reference Proteome 2021_03
## 68 leishmania 5664 LEIMA Reference Proteome 2021_03
## 69 leptospira 189518 LEPIN Reference Proteome 2021_03
## 70 listeria 169963 LISMO Reference Proteome 2021_03
## 71 macacque 9544 MACMU Reference Proteome 2021_03
## 72 cassava 3983 MANES Reference Proteome 2021_03
## 73 liverwort 3197 MARPO Reference Proteome 2021_03
## 74 barrel medic 3880 MEDTR Reference Proteome 2021_03
## 75 methanocaldococcus 243232 METJA Reference Proteome 2021_03
## 76 methanosarcina 188937 METAC Reference Proteome 2021_03
## 77 opossum 13616 MONDO Reference Proteome 2021_03
## 78 monosiga 81824 MONBE Reference Proteome 2021_03
## 79 banana 214687 MUSAM Reference Proteome 2021_03
## 80 mycobacterium 83332 MYCTU Reference Proteome 2021_03
## 81 meningococcus 122586 NEIMB Reference Proteome 2021_03
## 82 sacred lotus 4432 NELNU Reference Proteome 2021_03
## 83 nematostella 45351 NEMVE Reference Proteome 2021_03
## 84 aspergillus 330879 ASPFU Reference Proteome 2021_03
## 85 neurospora 367110 NEUCR Reference Proteome 2021_03
## 86 tobacco 4097 TOBAC Reference Proteome 2021_03
## 87 nitrosopumilu 436308 NITMS Reference Proteome 2021_03
## 88 platypus 9258 ORNAN Reference Proteome 2021_03
## 89 rice 39947 ORYSJ Reference Proteome 2021_03
## 90 oryzias 8090 ORYLA Reference Proteome 2021_03
## 91 chimpanzee 9598 PANTR Reference Proteome 2021_03
## 92 paramecium 5888 PARTE Reference Proteome 2021_03
## 93 phaeosphaeria 321614 PHANO Reference Proteome 2021_03
## 94 moss 3218 PHYPA Reference Proteome 2021_03
## 95 phytophthora 164328 PHYRM Reference Proteome 2021_03
## 96 plasmodium 36329 PLAF7 Reference Proteome 2021_03
## 97 black_cottonwood 3694 POPTR Reference Proteome 2021_03
## 98 pristionchus 54126 PRIPA Reference Proteome 2021_03
## 99 peach 3760 PRUPE Reference Proteome 2021_03
## 100 pseudomonas 208964 PSEAE Reference Proteome 2021_03
## 101 puccinia 418459 PUCGT Reference Proteome 2021_03
## 102 pyrobaculum 178306 PYRAE Reference Proteome 2021_03
## 103 rhodopirellula 243090 RHOBA Reference Proteome 2021_03
## 104 bean 3988 RICCO Reference Proteome 2021_03
## 105 salmonella 99287 SALTY Reference Proteome 2021_03
## 106 s_japonicus 402676 SCHJY Reference Proteome 2021_03
## 107 sclerotinia 665079 SCLS1 Reference Proteome 2021_03
## 108 spikemoss 88036 SELML Reference Proteome 2021_03
## 109 millet 4555 SETIT Reference Proteome 2021_03
## 110 shewanella 211586 SHEON Reference Proteome 2021_03
## 111 tomato 4081 SOLLC Reference Proteome 2021_03
## 112 potato 4113 SOLTU Reference Proteome 2021_03
## 113 sorghum 4558 SORBI Reference Proteome 2021_03
## 114 spinach 3562 SPIOL Reference Proteome 2021_03
## 115 staph 93061 STAA8 Reference Proteome 2021_03
## 116 strep 171101 STRR6 Reference Proteome 2021_03
## 117 streptomyces 100226 STRCO Reference Proteome 2021_03
## 118 sea_urchin 7668 STRPU Reference Proteome 2021_03
## 119 sulfolobus 273057 SACS2 Reference Proteome 2021_03
## 120 pig 9823 PIG Reference Proteome 2021_03
## 121 synechocystis 1111708 SYNY3 Reference Proteome 2021_03
## 122 thalassiosira 35128 THAPS Reference Proteome 2021_03
## 123 cacao 3641 THECC Reference Proteome 2021_03
## 124 thermococcus 69014 THEKO Reference Proteome 2021_03
## 125 thermodesulfovibrio 289376 THEYD Reference Proteome 2021_03
## 126 thermotoga 243274 THEMA Reference Proteome 2021_03
## 127 tribolium 7070 TRICA Reference Proteome 2021_03
## 128 trichomonas 5722 TRIVA Reference Proteome 2021_03
## 129 trichoplax 10228 TRIAD Reference Proteome 2021_03
## 130 wheat 4565 WHEAT Reference Proteome 2021_03
## 131 t_brucei 185431 TRYB2 Reference Proteome 2021_03
## 132 ustilago 237631 USTMA Reference Proteome 2021_03
## 133 cholera 243277 VIBCH Reference Proteome 2021_03
## 134 grape 29760 VITVI Reference Proteome 2021_03
## 135 xanthomonas 190485 XANCP Reference Proteome 2021_03
## 136 frog 8364 XENTR Reference Proteome 2021_03
## 137 yarrowia 284591 YARLI Reference Proteome 2021_03
## 138 yersinia 632 YERPE Reference Proteome 2021_03
## 139 maize 4577 MAIZE Reference Proteome 2021_03
## 140 eelgrass 29655 ZOSMR Reference Proteome 2021_03
## 141 helobdella 6412 HELRO Reference Proteome 2021_03
## 142 lepisosteudae 7918 LEPOC Reference Proteome 2021_03
## 143 m_genitalium 243273 MYCGE Reference Proteome 2021_03
## long_name
## 1 Homo sapiens
## 2 Mus musculus
## 3 Rattus norvegicus
## 4 Gallus gallus
## 5 Danio rerio
## 6 Drosophila melanogaster
## 7 Caenorhabditis elegans
## 8 Saccharomyces cerevisiae
## 9 Schizosaccharomyces pombe
## 10 Dictyostelium discoideum
## 11 Arabidopsis thaliana
## 12 Escherichia coli
## 13 Emericella nidulans
## 14 Amborella trichopoda
## 15 Anolis carolinensis
## 16 Anopheles gambiae
## 17 Aquifex aeolicus
## 18 Ashbya gossypii
## 19 Bacillus cereus
## 20 Bacillus subtilis
## 21 Bacteroides thetaiotaomicron
## 22 Batrachochytrium dendrobatidis
## 23 Bos taurus
## 24 Brachypodium distachyon
## 25 Bradyrhizobium diazoefficiens
## 26 Branchiostoma floridae
## 27 Brassica napus
## 28 Brassica rapa subsp. pekinensis
## 29 Caenorhabditis briggsae
## 30 Candida albicans
## 31 Canis lupus familiaris
## 32 Capsicum annuum
## 33 Chlamydia trachomatis
## 34 Chlamydomonas reinhardtii
## 35 Chloroflexus aurantiacus
## 36 Ciona intestinalis
## 37 Citrus sinensis
## 38 Clostridium botulinum
## 39 Coxiella burnetii
## 40 Cryptococcus neoformans
## 41 Cucumis sativus
## 42 Daphnia pulex
## 43 Deinococcus radiodurans
## 44 Dictyoglomus turgidum
## 45 Dictyostelium purpureum
## 46 Entamoeba histolytica
## 47 Equus caballus
## 48 Erythranthe guttata
## 49 Eucalyptus grandis
## 50 Felis catus
## 51 Fusobacterium nucleatum
## 52 Geobacter sulfurreducens
## 53 Giardia intestinalis
## 54 Gloeobacter violaceus
## 55 Glycine max
## 56 Gorilla gorilla gorilla
## 57 Gossypium hirsutum
## 58 Haemophilus influenzae
## 59 Halobacterium salinarum
## 60 Helianthus annuus
## 61 Helicobacter pylori
## 62 Hordeum vulgare subsp. vulgare
## 63 Ixodes scapularis
## 64 Juglans regia
## 65 Klebsormidium nitens
## 66 Korarchaeum cryptofilum
## 67 Lactuca sativa
## 68 Leishmania major
## 69 Leptospira interrogans
## 70 Listeria monocytogenes
## 71 Macaca mulatta
## 72 Manihot esculenta
## 73 Marchantia polymorpha
## 74 Medicago truncatula
## 75 Methanocaldococcus jannaschii
## 76 Methanosarcina acetivorans
## 77 Monodelphis domestica
## 78 Monosiga brevicollis
## 79 Musa acuminata subsp. malaccensis
## 80 Mycobacterium tuberculosis
## 81 Neisseria meningitidis serogroup b
## 82 Nelumbo nucifera
## 83 Nematostella vectensis
## 84 Neosartorya fumigata
## 85 Neurospora crassa
## 86 Nicotiana tabacum
## 87 Nitrosopumilus maritimus
## 88 Ornithorhynchus anatinus
## 89 Oryza sativa
## 90 Oryzias latipes
## 91 Pan troglodytes
## 92 Paramecium tetraurelia
## 93 Phaeosphaeria nodorum
## 94 Physcomitrella patens
## 95 Phytophthora ramorum
## 96 Plasmodium falciparum
## 97 Populus trichocarpa
## 98 Pristionchus pacificus
## 99 Prunus persica
## 100 Pseudomonas aeruginosa
## 101 Puccinia graminis
## 102 Pyrobaculum aerophilum
## 103 Rhodopirellula baltica
## 104 Ricinus communis
## 105 Salmonella typhimurium
## 106 Schizosaccharomyces japonicus
## 107 Sclerotinia sclerotiorum
## 108 Selaginella moellendorffii
## 109 Setaria italica
## 110 Shewanella oneidensis
## 111 Solanum lycopersicum
## 112 Solanum tuberosum
## 113 Sorghum bicolor
## 114 Spinacia oleracea
## 115 Staphylococcus aureus
## 116 Streptococcus pneumoniae
## 117 Streptomyces coelicolor
## 118 Strongylocentrotus purpuratus
## 119 Sulfolobus solfataricus
## 120 Sus scrofa
## 121 Synechocystis
## 122 Thalassiosira pseudonana
## 123 Theobroma cacao
## 124 Thermococcus kodakaraensis
## 125 Thermodesulfovibrio yellowstonii
## 126 Thermotoga maritima
## 127 Tribolium castaneum
## 128 Trichomonas vaginalis
## 129 Trichoplax adhaerens
## 130 Triticum aestivum
## 131 Trypanosoma brucei
## 132 Ustilago maydis
## 133 Vibrio cholerae
## 134 Vitis vinifera
## 135 Xanthomonas campestris
## 136 Xenopus tropicalis
## 137 Yarrowia lipolytica
## 138 Yersinia pestis
## 139 Zea mays
## 140 Zostera marina
## 141 helobdella robusta
## 142 lepisosteus oculatus
## 143 mycoplasma genitaliumFisher’s exact test with FDR < 0.01, GO Biological Process Complete annotation.
res_up <- rba_panther_enrich(genes = up_genes,
organism = 7955 , annot_dataset = "GO:0008150",
cutoff = 0.01,
test_type = "FISHER",
correction = "FDR")## Performing over-representation enrichment analysis of 406 input genes of organism 7955 against GO:0008150 datasets.res_down <- rba_panther_enrich(genes = down_genes,
organism = 7955 , annot_dataset = "GO:0008150",
cutoff = 0.01,
test_type = "FISHER",
correction = "FDR")## Performing over-representation enrichment analysis of 505 input genes of organism 7955 against GO:0008150 datasets.Plot results as diverging barcharts.
res_up <- data.frame(res_up$result)
res_up$GO <- paste(res_up$term.id,res_up$term.label,sep="_")
res_up$representation <- ifelse(res_up$fold_enrichment < 1, "under", "over")
res_up <- res_up[order(-res_up$fold_enrichment), ]
# convert to factor to retain sorted order in plot
res_up$GO <- factor(res_up$GO, levels = res_up$GO)
res_up <- res_up %>% filter(fold_enrichment > 10)
ggplot(res_up, aes(x=GO, y=fold_enrichment, label=fold_enrichment)) +
geom_bar(stat='identity', aes(fill=representation), width=.5) +
scale_fill_manual(name="Representation",
labels = c("Over", "Under"),
values = c("over"="#0072B2", "under"="#D55E00")) +
labs(subtitle="PANTHER statistical overrepresentation test",
title= "Upregulated genes qval < 0.05. Fisher's exact, FDR < 0.01, fold_enrichment > 10") +
coord_flip()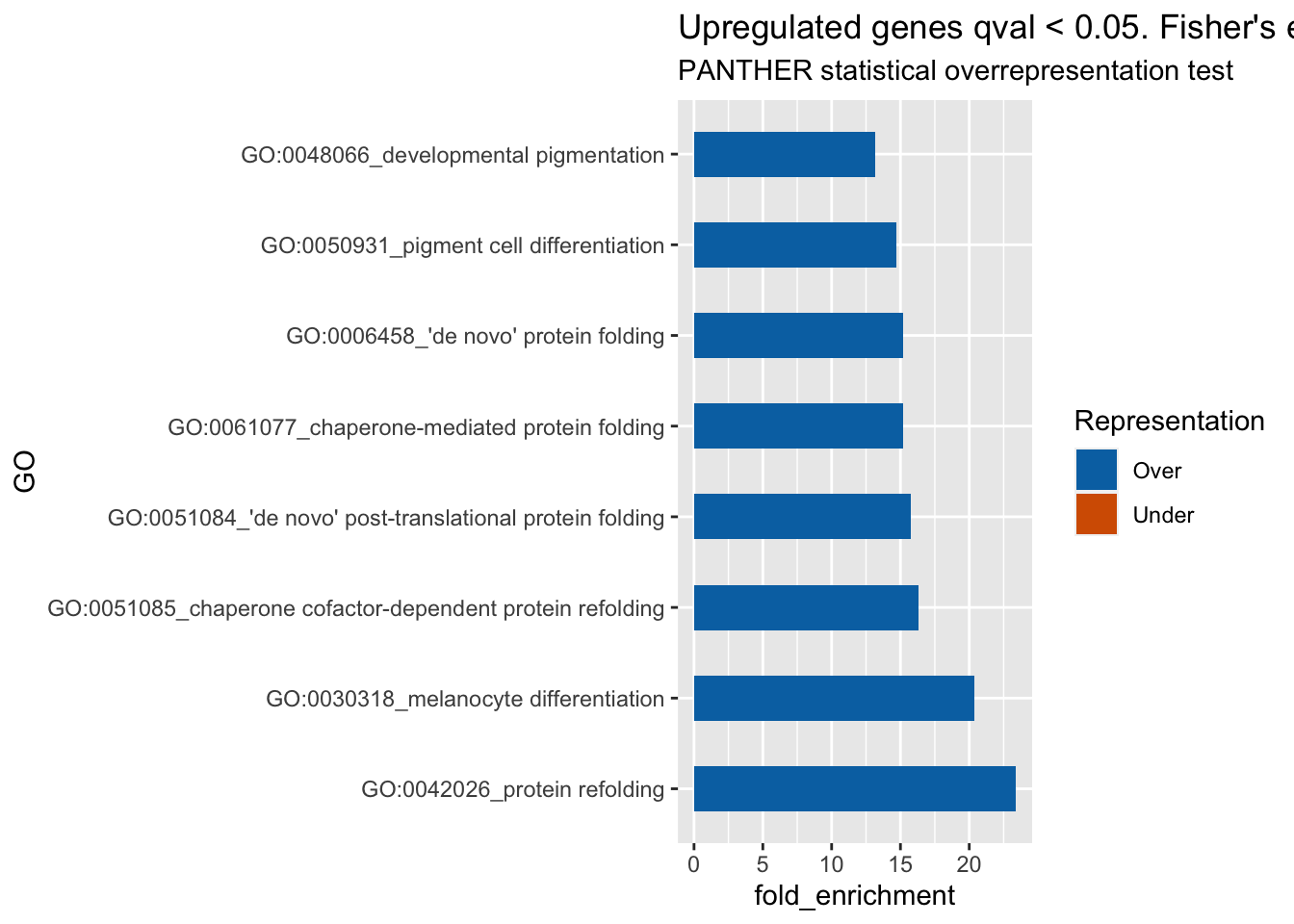
res_down <- data.frame(res_down$result)
res_down$GO <- paste(res_down$term.id,res_down$term.label,sep="_")
res_down$representation <- ifelse(res_down$fold_enrichment < 1, "under", "over")
res_down <- res_down[order(-res_down$fold_enrichment), ]
# convert to factor to retain sorted order in plot
res_down$GO <- factor(res_down$GO, levels = res_down$GO)
res_down <- res_down %>% filter(fold_enrichment > 10)
ggplot(res_down, aes(x=GO, y=fold_enrichment, label=fold_enrichment)) +
geom_bar(stat='identity', aes(fill=representation), width=.5) +
scale_fill_manual(name="Representation",
labels = c("Over", "Under"),
values = c("over"="#0072B2", "under"="#D55E00")) +
labs(subtitle="PANTHER statistical overrepresentation test",
title= "Downregulated genes qval < 0.05. Fisher's exact, FDR < 0.01, fold_enrichment > 10") +
coord_flip()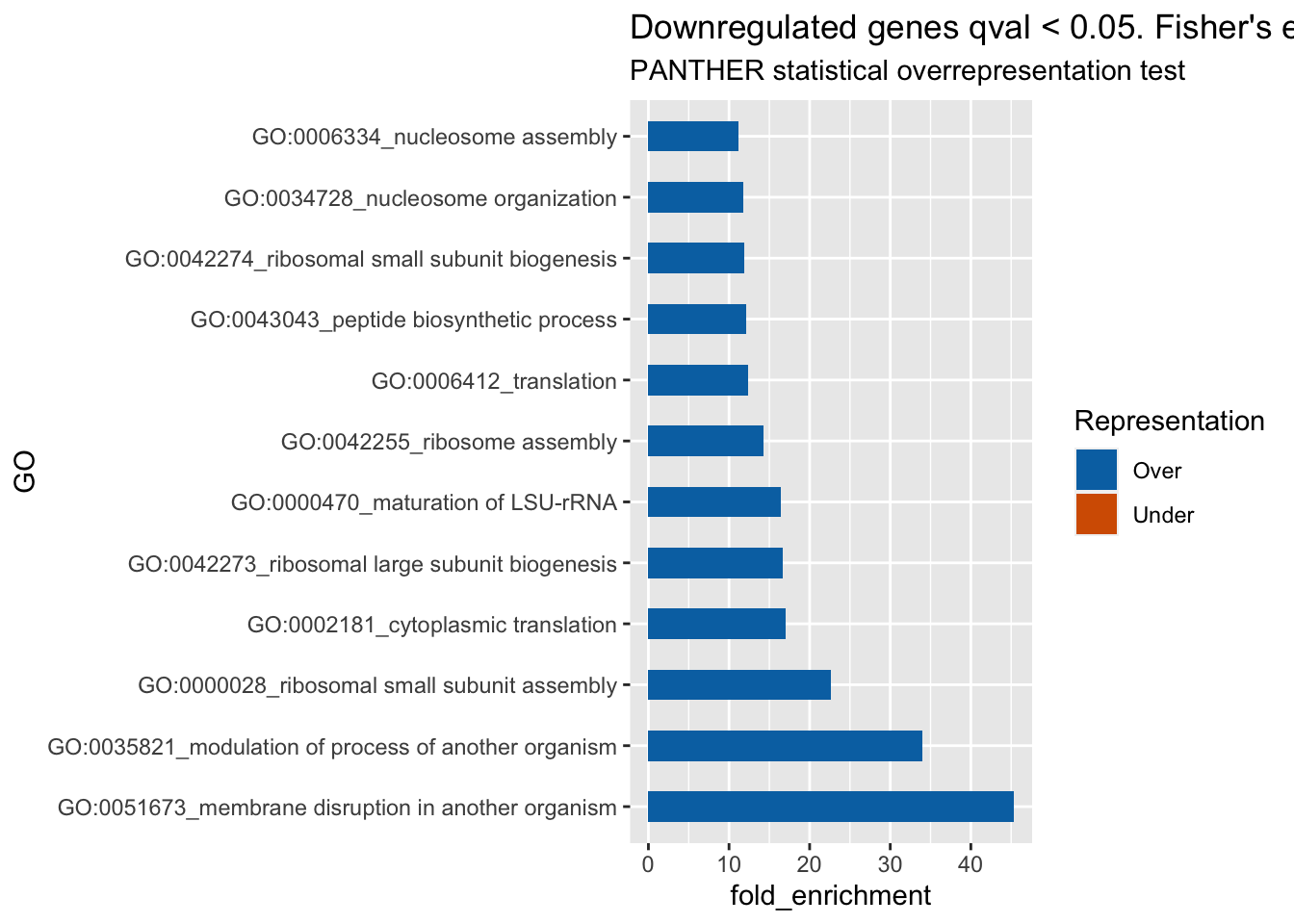
sessionInfo()## R version 4.2.1 (2022-06-23)
## Platform: x86_64-apple-darwin17.0 (64-bit)
## Running under: macOS Mojave 10.14.6
##
## Matrix products: default
## BLAS: /Library/Frameworks/R.framework/Versions/4.2/Resources/lib/libRblas.0.dylib
## LAPACK: /Library/Frameworks/R.framework/Versions/4.2/Resources/lib/libRlapack.dylib
##
## locale:
## [1] en_GB.UTF-8/en_GB.UTF-8/en_GB.UTF-8/C/en_GB.UTF-8/en_GB.UTF-8
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] rbioapi_0.7.7 biomaRt_2.52.0 forcats_0.5.2 stringr_1.4.1
## [5] dplyr_1.0.10 purrr_0.3.5 readr_2.1.3 tidyr_1.2.1
## [9] tibble_3.1.8 ggplot2_3.3.6 tidyverse_1.3.2 sleuth_0.30.0
##
## loaded via a namespace (and not attached):
## [1] matrixStats_0.62.0 bitops_1.0-7 fs_1.5.2
## [4] lubridate_1.8.0 bit64_4.0.5 RColorBrewer_1.1-3
## [7] filelock_1.0.2 progress_1.2.2 httr_1.4.4
## [10] GenomeInfoDb_1.32.4 tools_4.2.1 backports_1.4.1
## [13] bslib_0.4.0 utf8_1.2.2 R6_2.5.1
## [16] DBI_1.1.3 lazyeval_0.2.2 BiocGenerics_0.42.0
## [19] colorspace_2.0-3 rhdf5filters_1.8.0 withr_2.5.0
## [22] gridExtra_2.3 prettyunits_1.1.1 tidyselect_1.2.0
## [25] curl_4.3.3 bit_4.0.4 compiler_4.2.1
## [28] cli_3.4.1 rvest_1.0.3 Biobase_2.56.0
## [31] xml2_1.3.3 labeling_0.4.2 sass_0.4.2
## [34] scales_1.2.1 rappdirs_0.3.3 digest_0.6.30
## [37] rmarkdown_2.17 XVector_0.36.0 pkgconfig_2.0.3
## [40] htmltools_0.5.3 highr_0.9 dbplyr_2.2.1
## [43] fastmap_1.1.0 rlang_1.0.6 readxl_1.4.1
## [46] rstudioapi_0.14 RSQLite_2.2.18 farver_2.1.1
## [49] jquerylib_0.1.4 generics_0.1.3 jsonlite_1.8.3
## [52] googlesheets4_1.0.1 RCurl_1.98-1.9 magrittr_2.0.3
## [55] GenomeInfoDbData_1.2.8 Rcpp_1.0.9 munsell_0.5.0
## [58] Rhdf5lib_1.18.2 S4Vectors_0.34.0 fansi_1.0.3
## [61] lifecycle_1.0.3 stringi_1.7.8 yaml_2.3.6
## [64] zlibbioc_1.42.0 plyr_1.8.7 BiocFileCache_2.4.0
## [67] rhdf5_2.40.0 grid_4.2.1 blob_1.2.3
## [70] parallel_4.2.1 crayon_1.5.2 Biostrings_2.64.1
## [73] haven_2.5.1 hms_1.1.2 KEGGREST_1.36.3
## [76] knitr_1.40 pillar_1.8.1 reshape2_1.4.4
## [79] stats4_4.2.1 reprex_2.0.2 XML_3.99-0.11
## [82] glue_1.6.2 evaluate_0.17 data.table_1.14.4
## [85] modelr_0.1.9 vctrs_0.5.0 png_0.1-7
## [88] tzdb_0.3.0 cellranger_1.1.0 gtable_0.3.1
## [91] assertthat_0.2.1 cachem_1.0.6 xfun_0.34
## [94] broom_1.0.1 googledrive_2.0.0 gargle_1.2.1
## [97] pheatmap_1.0.12 AnnotationDbi_1.58.0 memoise_2.0.1
## [100] IRanges_2.30.1 ellipsis_0.3.2